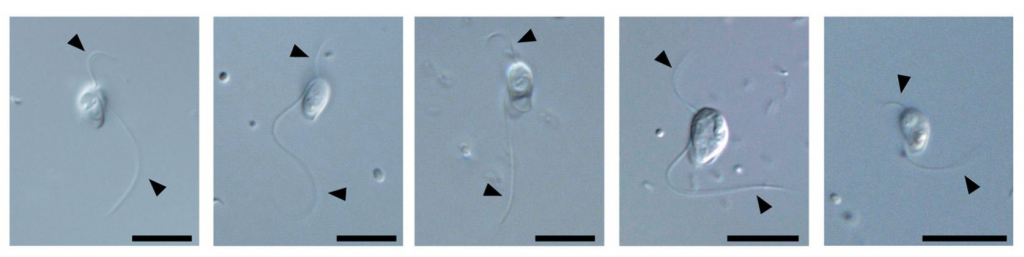
吉永真理(Yoshinaga Mari)さん
理工情報生命学術院 生命地球科学研究群
生物学学位プログラム
稲垣研究室(微生物分子進化研究室)博士前期課程1年
(内容は、2020年8月取材当時のものです。)
吉永さんは、筑波大学 生命環境学群 生物学類在籍時に微生物分子進化研究室に入り、稲垣祐司教授の指導のもとで研究を続けています。
今の研究室を選んだ理由
3年生のときに、「分子進化学」という稲垣先生と橋本先生の講義を受けて、とても興味を持ちました。
高校までは、生物というと、マウスやハエなどの生き物を育てたり、実験したりするイメージがとても強かったのですが、稲垣先生たちの講義では、パソコンを使って統計学的な解析をするということで、とても新鮮でした。
それまで、生物の分野でそんな研究ができることも知らなかったので、自分でやってみたいと思いました。
どんな研究をしているの?
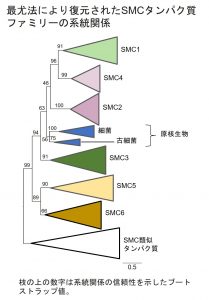
卒業研究で行っていた「SMC遺伝子の多様化と二次的喪失」というテーマを続けています。SMC は、染色体構造維持(Structural Maintenance of Chromosomes)の略です。SMCタンパク質は色々な働きを持っています。たとえば、生物が細胞分裂を行うときは、染色体を均等に分ける必要があるのですが、このとき染色体をバラバラにせずコンパクトに圧縮する機能や、複製された二本の姉妹染色分体がバラバラにならないように一緒に束ねておく機能を持っています。
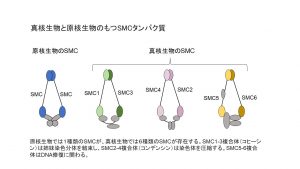
とても重要な機能を担っているので、あらゆる生物が持っていると言われているのですが、本当にすべての生物がSMCタンパク質を持っているのか、そしてその系統について大規模に解析された事例はなかったので、今回私の研究で初めて大規模解析を行いました。
今回推測した系統樹から、真核生物のSMCは単に原核生物(細菌、古細菌)のSMCから進化したのではなく、もっと複雑な進化過程を経て多様化したのではないか、と考察できました。
今は、研究成果を論文として投稿する準備をしています。
どんな風に研究をしているの?
まずは色々な種のSMCタンパク質に関する配列を集めてきて、データセットを作ります。データセットをそろえたら、系統解析を行います。
解析には、研究室にある計算機を使います。系統解析の場合、早ければ2−3日で終わりますが、解析に使うモデルが複雑になると1ヶ月近くかかる計算もあります。

パソコンでの操作は、研究室に入るまでは慣れていなくて、研究室に入ってから勉強したのですが、研究室の先輩が丁寧に教えてくれました。

CCSで研究するまでにどんな勉強をしたの?
高校の理科は物理と化学を選択していたので、生物はやっていませんでした。でも、生命の不思議について興味があったので、生物の研究環境が整っている筑波大学を受験して入学しました。大学に入ってからは、生物の講義や実習があって、慣れるまでは大変でしたが、周りの人も優しく教えてくれるので、大丈夫でした。
メッセージ
分子進化学は、生物の一般的なイメージとは違って、コンピュータを使った統計解析などをします。生物の分野でも、こんな研究があるということを皆さんにぜひ知ってもらいたいです。
【CCSで学ぶ】一覧に戻る