Group Leader
 |
INAGAKI Yuji, Professor
Doctor Yuji Inagaki began his professional career as a Postdoctoral Fellow in the Program of Evolutionary Biology at the Canadian Institute for Advanced Research in 2000. In 2004, he became a lecturer at the Nagahama Institute for Bioscience and Technology before moving to University of Tsukuba as an Assistant Professor in 2005. Dr. Yuji Inagaki was awarded the Young Scientists Prize by the Ministry of Education, Culture, Sports, Science and Technology (MEXT) in 2007, and received the Young Scientist Initiative Award from the Society of Evolutionary Studies, Japan in 2008. |
Overview
Building the global phylogeny of eukaryotes
All living organisms on Earth are believed to have evolved from a single common ancestral cell, eventually diverging into the three “domains,” of Bacteria, Archea (Archaebacteria), and Eukarya. Needless to say, biological evolution has never stopped, and has resulted in the creation of countless numbers of living organisms. One of the most important and difficult fields of biological study involves the evolutionary relationship amongst all the life forms on earth – building the “Universal Tree of Life”.
The central focus of our research is working to gain insights into the origin and early evolution of eukaryotes (members of the Eukarya domain). From the results of pioneering studies conducted to date, it is now believed that the vast majority of eukaryotes belong to any of ten major assemblages. One of the goals of this research is reconstructing the relationship amongst the major taxonomic groups in Eukarya, and this issue has been tackled by using molecular phylogenetic techniques.
Research Topics
Analyzing ‘phylogenomic’ data comprising >100 gene sequences
Until recently, phylogenetic trees were constructed from the information of a single gene or a few genes. However, the information of a single gene is insufficient to make robust phylogenetic inferences related to ancient events such as the separations of major eukaryotic groups. To counter the difficulties of single-gene analyses, sequence data comprised of more than 100 genes have taken center stage in more recent phylogenetic analyses. These ‘phylogenomic’ analyses have provided successful insights into the relationship between major eukaryotic assemblages.
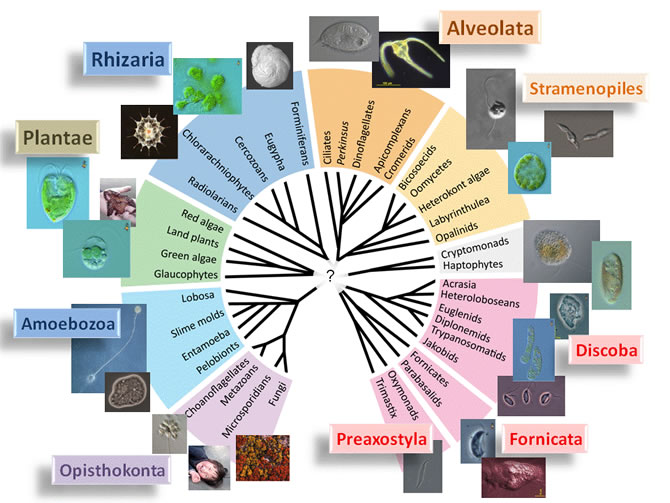 Fig.1 Tree of eukaryotes
Fig.1 Tree of eukaryotes
Current ongoing projects
We are specifically interested in several eukaryotic groups, such as cryptophytes (and their close relatives), Discoba, and Fornicata, as these may hold the keys to understanding eukaryotic cell evolution, particularly the evolutions of two important organelles – mitochondria and plastids (or chloroplasts). Cryptophytes are photosynthetic eukaryotes bearing plastids, but recent studies have indicated that cryptophytes and various non-photosynthetic lineages form a previously identified monophyletic group. Thus, the group represented by cryptophytes can be used as a model group to study the birth and death of plastids. Discoba is known as the group containing a species with the most ancestral mitochondrion, while many members of the Fornicata group lack classical mitochondria. We expect that these two groups will provide important insights into study of mitochondria evolution. Besides previously described eukaryotes, there are potentially many eukaryotic species/lineages remain unstudied in the natural environments. As those “novel eukaryotes” may provide clues for various issues in the evolution of eukaryotes, we are hunting them in the environments and subject them to phylogenomic analyses. Since a reliable phylogenetic inference simultaneously requires numerous genes and species, large-scale analyses using high performance computers play a key role in our study.
Web sites
Molecular Evolution of Microbes (Japanese)
(Update: Dec. 18, 2019)
