Chief
 |
SHIGETA Yasuteru, Professor Dr. Yasuteru Shigeta graduated from Department of Chemistry, Osaka University in 2000. Following his graduation, he initially served as a postdoctoral fellow of Japan Society for the Promotion Science working at the University of Tokyo. He served as an assistant professor in the University of Tokyo (2004), a Lecturer in University of Tsukuba (2007), an associate professor in University of Hyogo (2008), and an associate professor in Osaka University (2010). Since 2014, he joined the Center for Computational Sciences (CCS) in the University of Tsukuba as a full professor. |
Overview
Phenomena expressing in life are dominated by a series of chemical reactions driven by macromolecules such as proteins, nucleic acids, lipids, and sugars. Therefore, the fundamental molecular mechanisms of biological phenomena can be clarified by investigating the electronic structure change and the spatial arrangements of atoms accompanied by the chemical reactions. In our group, dynamic structure-function correlations of biomolecules are explored by using first-principles and classical molecular dynamics simulations.
Research topics
Structural transitions of proteins for understanding its function
We have developed an efficient sampling method to induce structural changes of proteins, which are extremely relevant to protein functions such as protein folding, domain motion, induced-fit, and protein complex formation processes. This method is a very suitable method for massively parallel computer environment.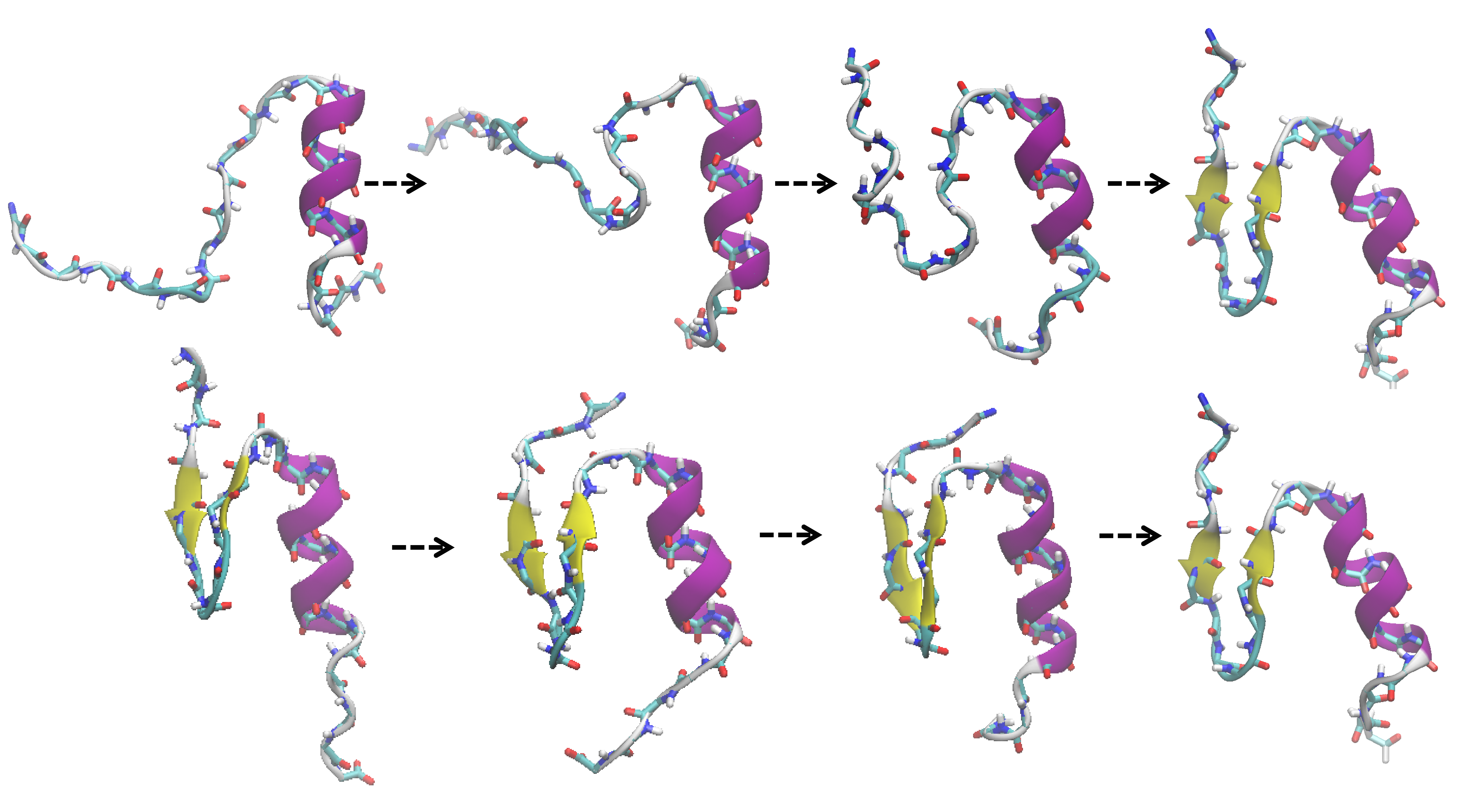
Enzymatic reaction analyses by using hybrid QM/MM calculations
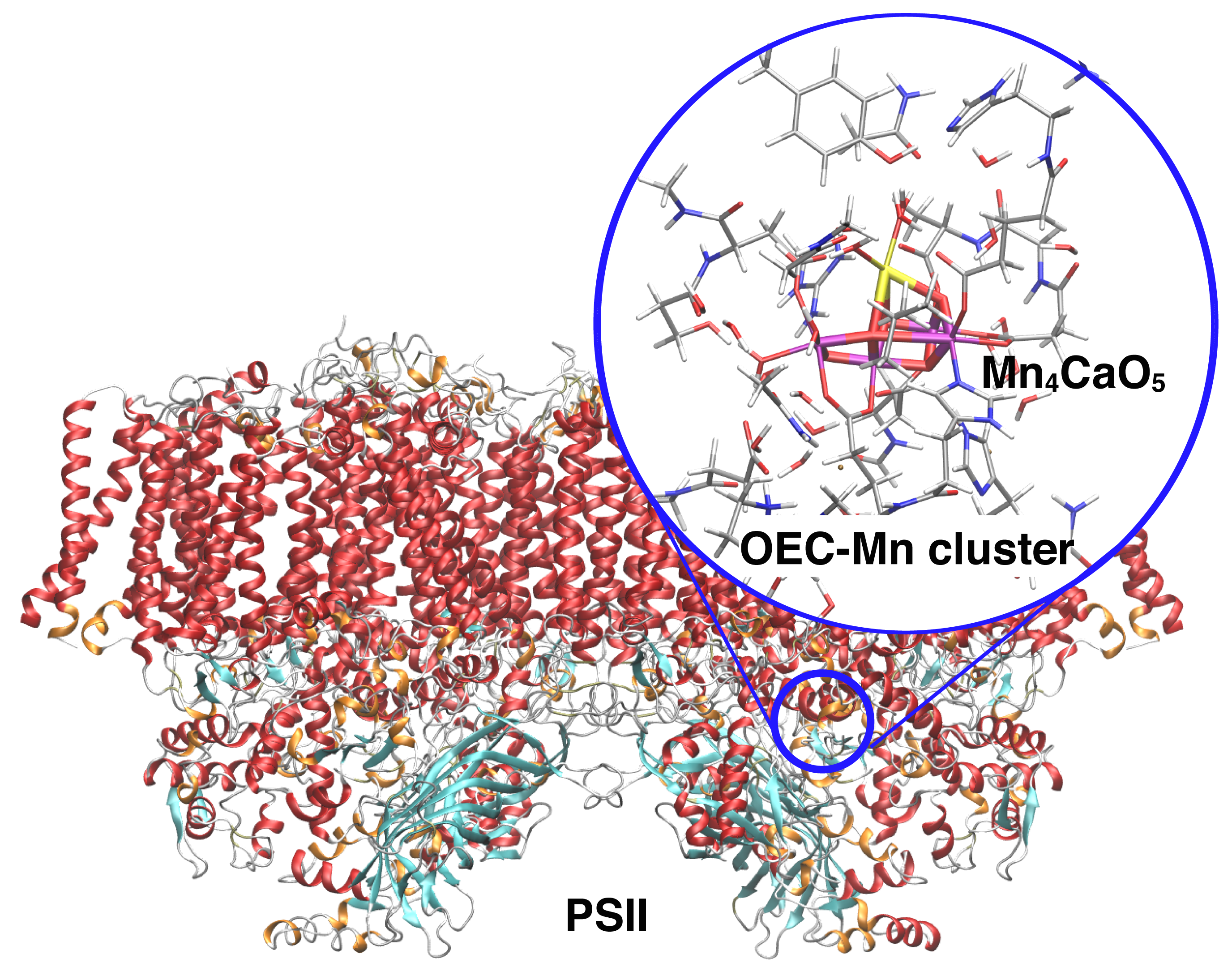
Photosystem II has a unique reaction active center composed of manganese, calcium, and oxygen atoms, where oxygen molecules are generated from water molecules by multi-step chemical reactions using light energy. We have clarified the reaction mechanisms of a series of water decomposition reactions by using the hybrid QM /MM method.
Origin of amino acids and its enantiomeric excess in space
Since some of the amino acids in life are also found in the meteorite, there is a possibility that an origin of life exists in the interstellar space. In collaboration with the Division of Astrophysics, our group has studied molecular evolution and symmetry breaking processes of the amino acids in space.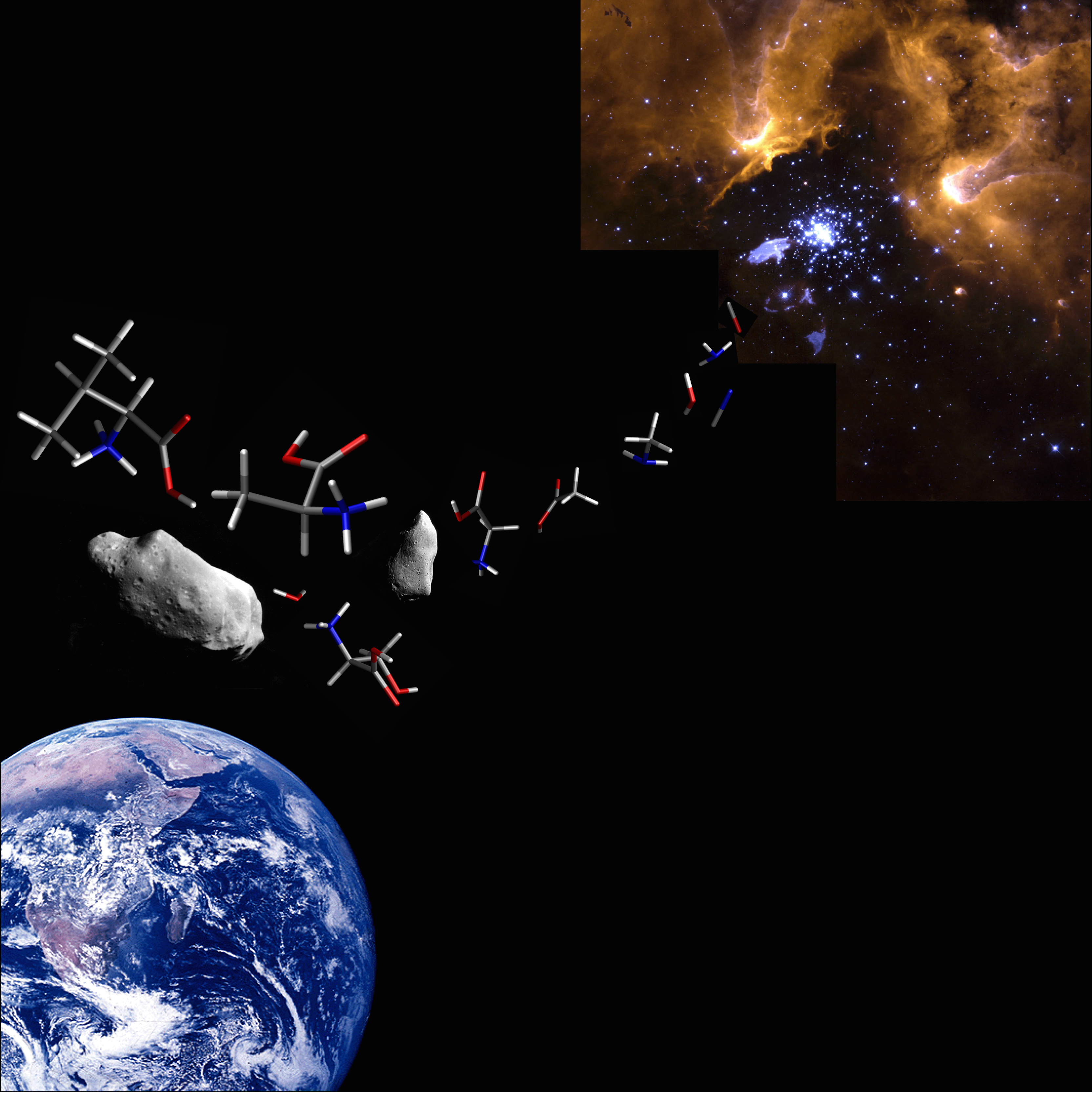
(Update: Dec. 18, 2019)